
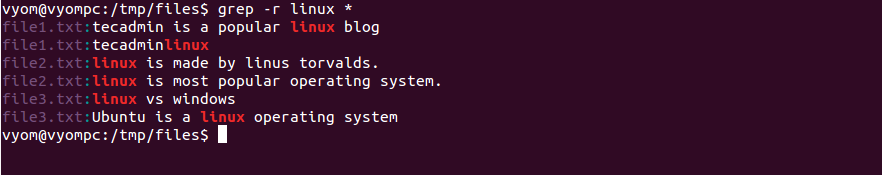
You may have noticed that the last two rows of the table show that Smith Island has 1 species in “Pine- hardwood_forest_stands”, and 40 species in “Pine- Hardwood_forest_stands”. # 8 Smith Pine-Hardwood_forest_stands 40Ĭool! This is useful information to know, and it’s all thanks to grepl() that we were able to perform this operation so easily. # 3 Parramore Pine-Hardwood_forest_stands 31 # View summary table print(forest_summary)
Here, I used grep() to subset the data to return all rows where a species is found in forested habitat.

Now that we know what grep() and grepl() return, we can subset our data frame using their outputs. grepl() returns TRUE when there is a match, and FALSE when there isn’t one, all the way down the entire data frame. The extra “l” in grepl() stands for “logical”, which is the data type that it returns. # FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE # FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE # FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE # TRUE TRUE FALSE TRUE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE # View the grepl output head(hab_log, 60) Hab_log <- grepl( "forest", veg_dat $habitat, ignore.case = TRUE) # Assign the list of values to a variable so we don't have to see all of them (it's a long list) If we add another argument to grep() that says value = TRUE, we can see the values where the function has found a match (in this case, the actual habitat types). You can see that grep() returns every row number in the data frame where a habitat type contains the string “forest”. # Let's see how grep works grep( "forest", veg_dat $habitat, ignore.case = TRUE) This tells the function that you don’t want your search to be case-sensitive (if you leave the ignore.case argument out, the default is that the function is case-sensitive). You can also add an argument to grep() where you set ignore.case = TRUE. You can use the function like so: grep(pattern_text, vector). Luckily, we have the grep() function to help us with that! The easiest way to pick all of these habitats out of the data set would be if we could “ctrl-F” the word “forest” in the habitat column. We also have “Pine-hardwood_forest_stands”, which is the same as the first one, but identified as a separate entry because “hardwood” is not capitalized. It looks like we have several types of forest: “Pine-Hardwood_forest_stands”, “Hardwood_forest_stands”, and “Pine_forest_stands”. # "Beachgrass_dunes-Dense_grassland_dunes" # View all unique values in the habitat column unique(veg_dat $habitat) How many habitat types do we have that are forested? We can use the unique() function to view all the unique entries for the habitat column. Let’s say that we’re interested in looking at all species that are found in forested habitats. We have a column for genus, species, island, habitat type, and the relative abundance of the species.

This data set lists observations of species presence on different islands and in different habitats on those islands. # 6 Achillea millefolium Smith Foredune_grassland 4 # 5 Achillea millefolium Smith Dense_grasslands 4 # 4 Achillea millefolium Smith Hardwood_forest_stands 4 # 3 Achillea millefolium Wreck Foredune_grassland 3 # 2 Acer rubrum Parramore Hardwood_forest_stands 6 # 1 Acer rubrum Smith Pine-Hardwood_forest_stands 6 Veg_dat <- dplyr :: select(veg_dat, genus, species, island, habitat, relabund)


 0 kommentar(er)
0 kommentar(er)
